The Cell Ontology
The Cell Ontology (CL) is an OBO Foundry ontology covering the domain of biological cell types with curation focused on animal cell types and interoperability with specialized ontologies of cell types in other areas of biology.


The Cell Ontology is built on FAIR principles and is tightly integrated with other ontologies including the Uberon multi-species anatomy ontology, which CL uses to record cell location and the Gene Ontology (GO) which uses CL as its main reference for cell types and which CL uses to record cell function .
The Cell Ontology actively engages with the community. Editors from multiple projects are embedded in the team, are active and responsive in discussions on the issue tracker, and work to improve the content and of the ontology and its technical infrastructure.
The Cell Ontology is also integrated into tools used by the community such as Ubergraph which allows logical queries, such as finding cell types by location and the Ontology Access Kit (OAK), and is uploaded into popular ontology browsers such as the Ontology Lookup Service (OLS) and Ontobee.
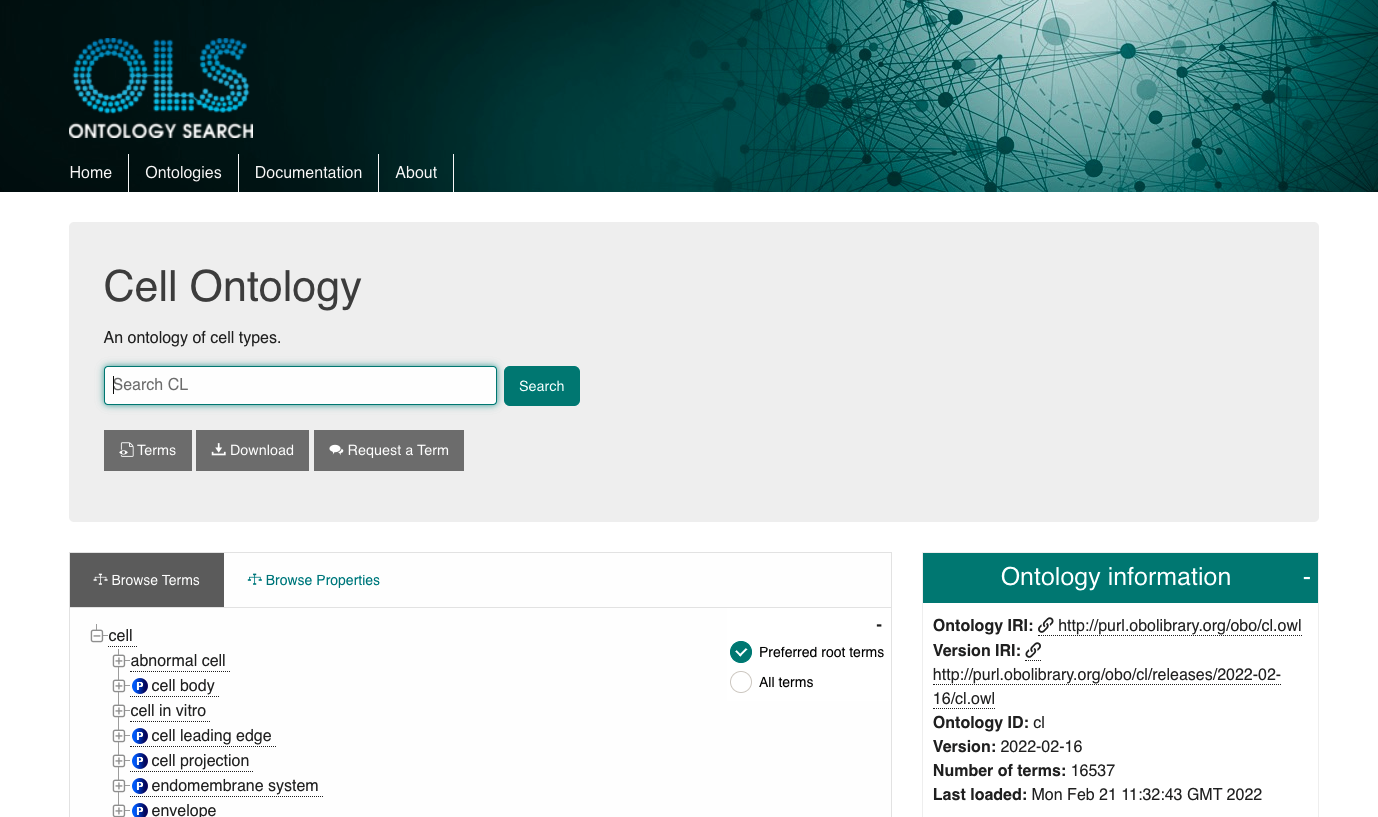
The Cell Ontology is released in multiple standard formats: owl (RDF/XML), obo, and json (obographs) format. It also comes in multiple variants including full (all imports merged in, classified using a reasoner), base (not pre-reasoned, only axioms belonging to the ontology, including those referencing non-CL classes), and simple (pre-reasoned, only CL classes), all of which are accessible from GitHub. All downloads come with a resolvable version IRI for persistent reference and access.
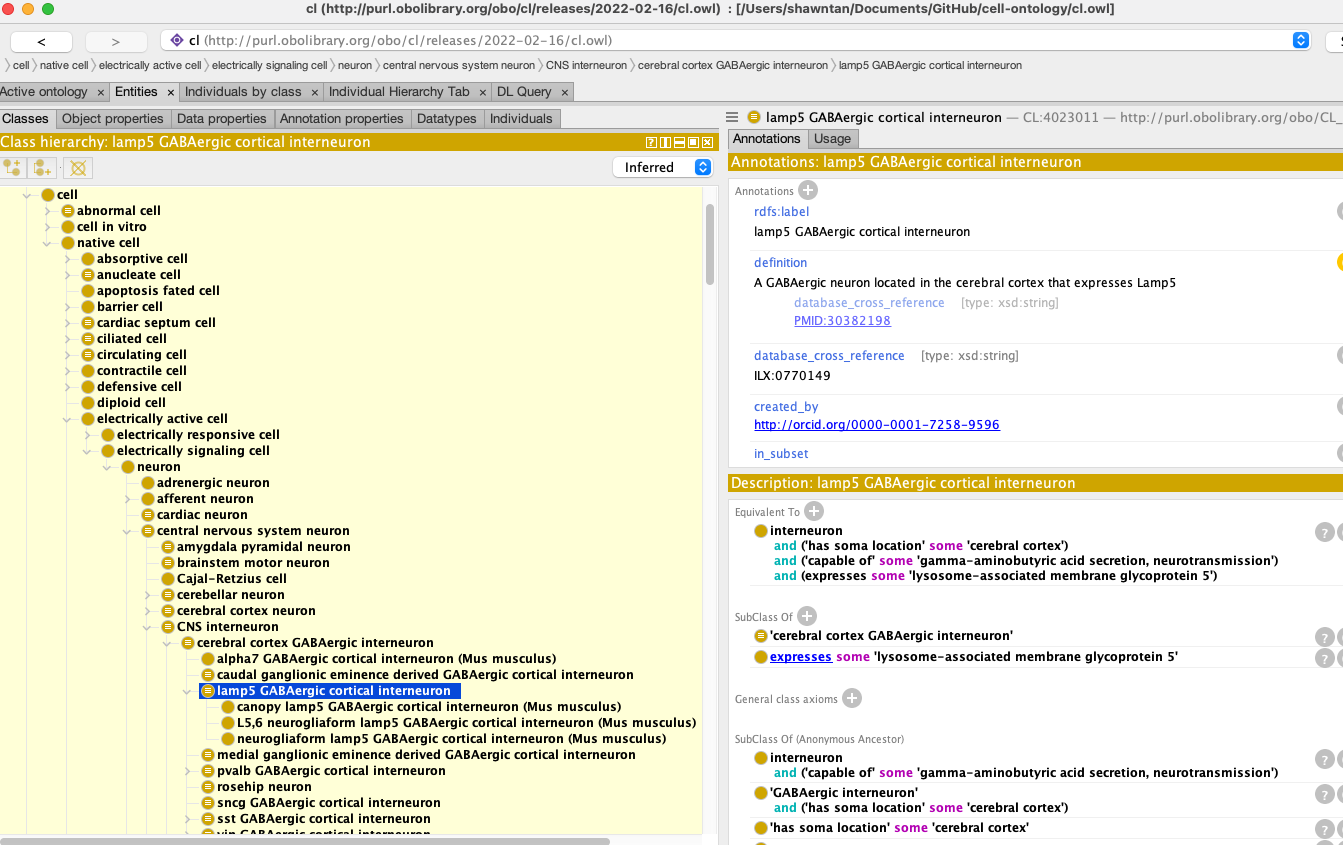
The Brain Data Standards Ontology (BDSO) is a data-driven extension of the Cell Ontology (CL) that supports the navigation, search, and organisation of information about cell types through an integrated web portal, and also functions as an independent ontology for use in cell-type annotation.
Here, we present OnClass, an algorithm and accompanying software for automatically classifying cells into cell types that are part of the controlled vocabulary that forms the Cell Ontology.
We worked with developers of external ontologies such as Uberon, the Cell Ontology, and the Fungal Anatomy Ontology to include taxon constraints within these ontologies, and to have these inherited in GO. For example, the Cell Ontology now constrains the term ‘leukocyte’ to vertebrates, and consequently terms in GO such as monocyte differentiation are inferred to be constrained to vertebrates.
The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability. Alexander D Diehl et al., J Biomed Semantics. 2016 Jul 4;7(1):44. PMID:27377652, PMCID:PMC4932724, DOI:10.1186/s13326-016-0088-7
Logical development of the cell ontology. Terrence F Meehan et al., BMC Bioinformatics. 2011 Jan 5;12:6. PMID:21208450, PMCID:PMC3024222, DOI:10.1186/1471-2105-12-6
Cell type discovery and representation in the era of high-content single cell phenotyping. Bakken, Trygve, et al., BMC Bioinformatics. Dec 21;18(Suppl 17):559. PMID: 29322913 PMCID: PMC5763450 DOI: 10.1186/s12859-017-1977-1
Hematopoietic cell types: prototype for a revised cell ontology. Diehl, Alexander D., et al., Journal of biomedical informatics. 2011 Feb 1;44(1):75-9. PMID: 20123131 PMCID: PMC2892030 DOI: 10.1016/j.jbi.2010.01.006
An improved ontological representation of dendritic cells as a paradigm for all cell types. Masci, Anna Maria, et al., BMC bioinformatics. 2009 Feb 25;10(1):70. PMID: 19243617 PMCID: PMC2662812 DOI: 10.1186/1471-2105-10-70
The preferred point of contact for the Cell Ontology is the GitHub Issue Tracker.